Publication on mutational processes and epigenetics in cancer
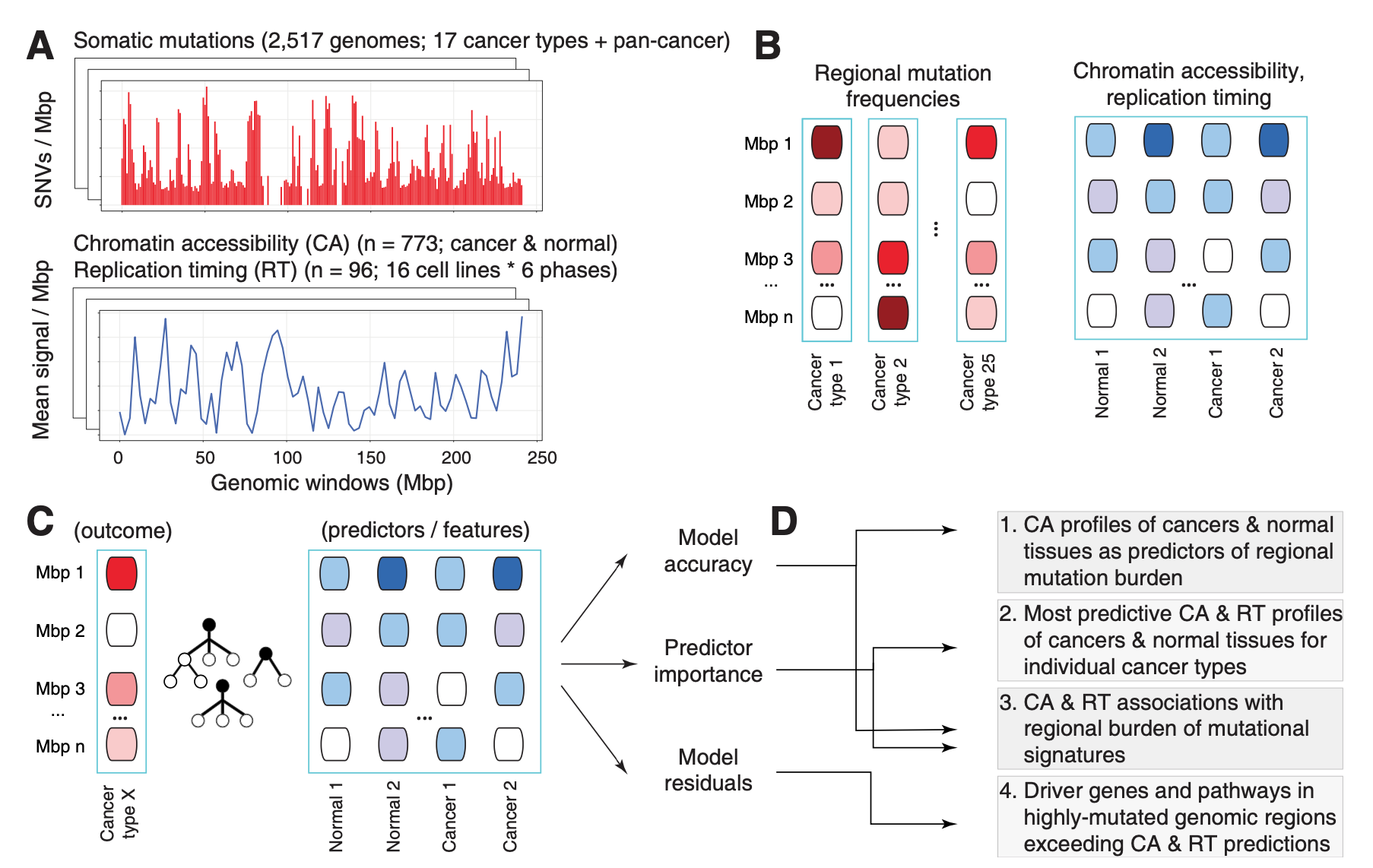
Our comprehensive analysis of mutational processes in cancer genomes and their epigenetic associations with tumor tissue of origin is now published in PLOS Computational Biology. Using a machine learning approach and a compendium of ~900 genome-wide epigenetic tracks of primary cancers and normal tissues, we predicted megabase-scale mutation rates in ~2400 whole cancer genomes and asked which epigenetic predictors contributed the most to the predictions of somatic mutation burden and mutational signatures.
We found that the epigenomes of primary cancers, rather than those of normal tissues, are the predominant predictors of somatic mutations in most cancer types. In the vast majority of case we found tissue-specific associations: for example, the distribution of somatic mutations in breast cancer genomes was most predictable by the chromatin measurements of primary human breast cancers and replication timing profiles of breast cancer cell lines, while normal breast tisses were not identified as predictors. This suggests that the mutational landscape of cancer genomes is more strongly determined by the cancer-specific chromatin environment that occurs after transformation of normal cells to cancer cells.
The analysis also revealed a landscape of epigenetic associations with mutational processes, where certain mutational signatures were more strongly driven by the epigenetic properties of the genome, especially carcinogenic signatures and signatures of unknown origin.
Lastly, we demonstrated that the epigenome-informed models can point out regions with known and putative driver mutations, as epigenomes explain a large fraction of the genomic variation and any regions with over-represented mutations exceeding these machine learning driven predictors.
The study was led by our graduate student Oliver Ocsenas of the Department of Medical Biophysics of University of Toronto who recently defended his Master’s thesis on the project.
(>) Chromatin accessibility of primary human cancers ties regional mutational processes and signatures with tissues of origin. Oliver Ocsenas, Jüri Reimand. (2022) PLOS Computational Biology e1010393. paper