Publication on human genetic variation of signaling networks of SARS-CoV-2 infection
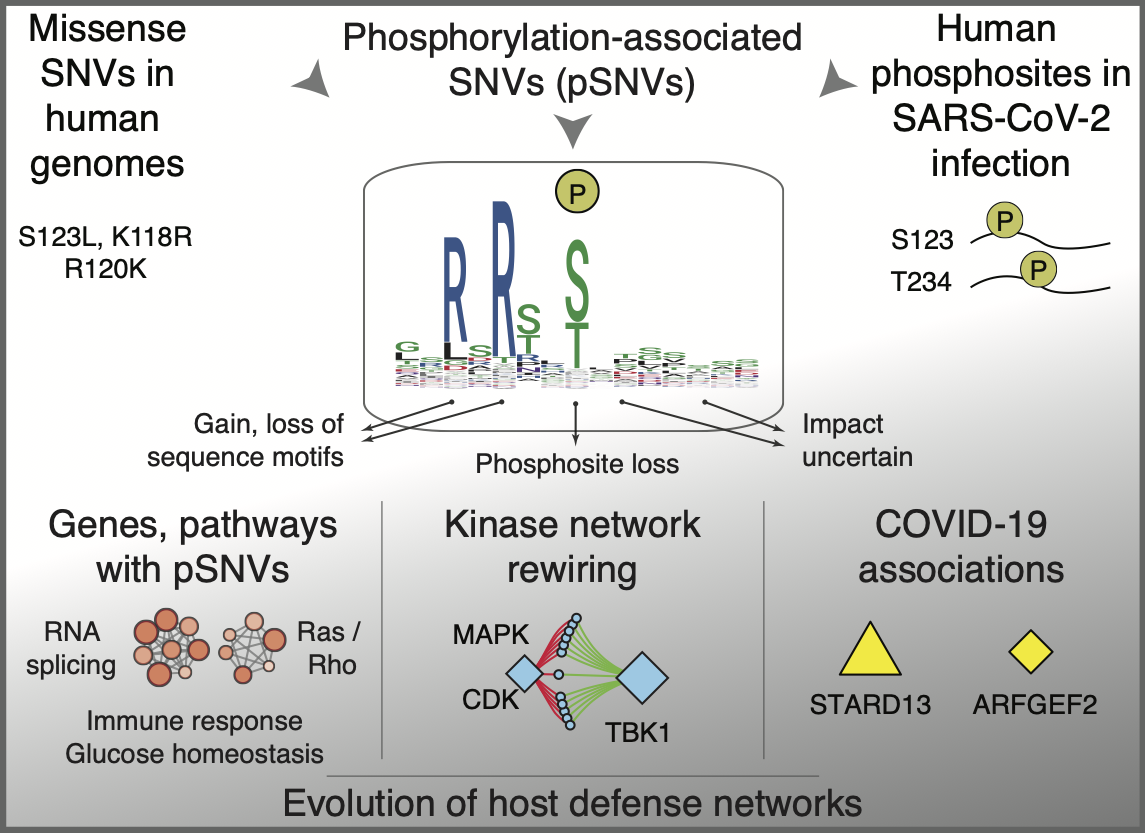
On our recent proteogenomics study of SARS-CoV-2 infection is now published in Molecular Systems Biology (1). We analysed human genetic variation affecting the phosphorylation signaling networks activated in virus infection using our previously developed machine learning approach (MIMP (2)). MIMP identified protein-coding variants (SNVs, single nucleotide variants) in human genomes that rewire signaling networks of SARS-CoV-2 infection by altering protein sequence motifs bound by kinases.
These phosphorylation-associated SNVs (pSNVs) are enriched in cellular pathways involved in virus life cycle such as alternative splicing, innate immune response, protein-protein interaction networks of host and virus proteins, as well as pathways involved in human comorbidities of COVID-19 (such as diabetes). We found several pSNVs that associate with severe COVID-19 disease risk and hospitalization, including one variant with a potential protective effect. A subset of pSNVs created binding sites of the TBK1 kinase involved in immune response by replacing motifs of mitotic kinases (MAPKs, CDKs), indicating evolution of defense responses in human genomes.
The study was co-led by two members our lab: Diogo Pellegrina, a postdoctoral researcher, and Alec Bahcheli, a graduate student of the Molecular Genetics Department of University of Toronto.
(1) Human phospho‐signaling networks of SARS‐CoV‐2 infection are rewired by population genetic variants. Diogo Pellegrina*, Alexander T Bahcheli*, Michal Krassowski, Jüri Reimand. (2022) Molecular Systems Biology 18 (5), e10823. paper
(2) MIMP: predicting the impact of mutations on kinase-substrate phosphorylation. Omar Wagih, Jüri Reimand*, Gary D Bader*. (2015) Nature Methods 12 (6), 531-533. paper github